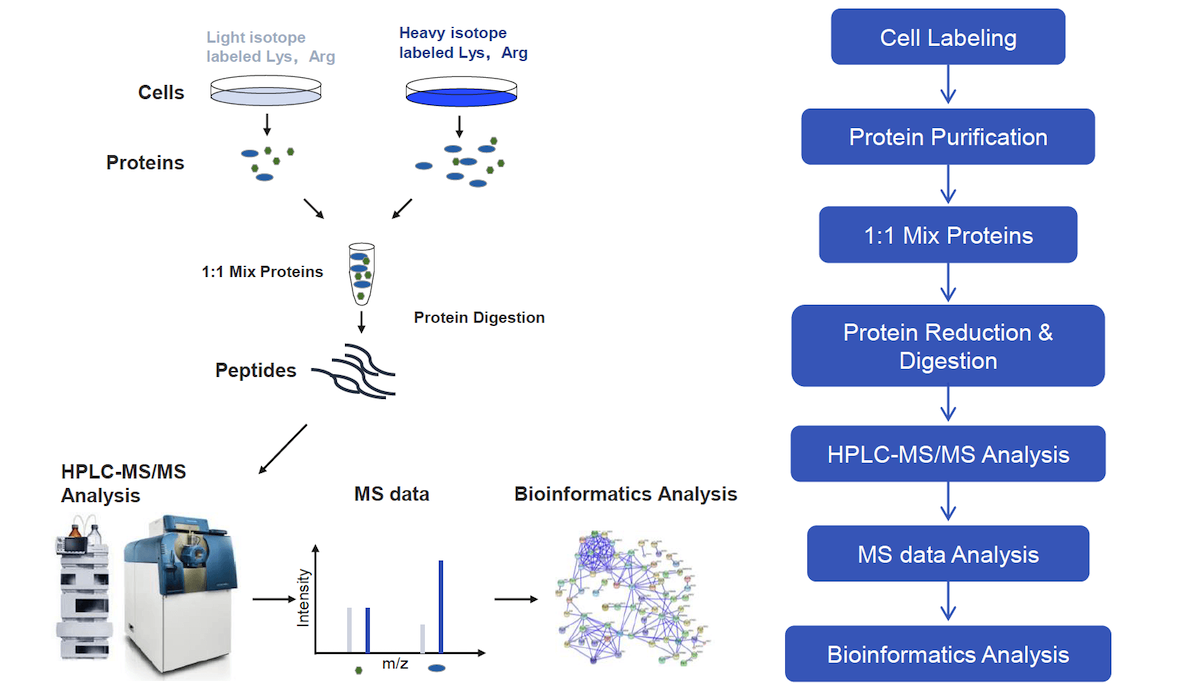
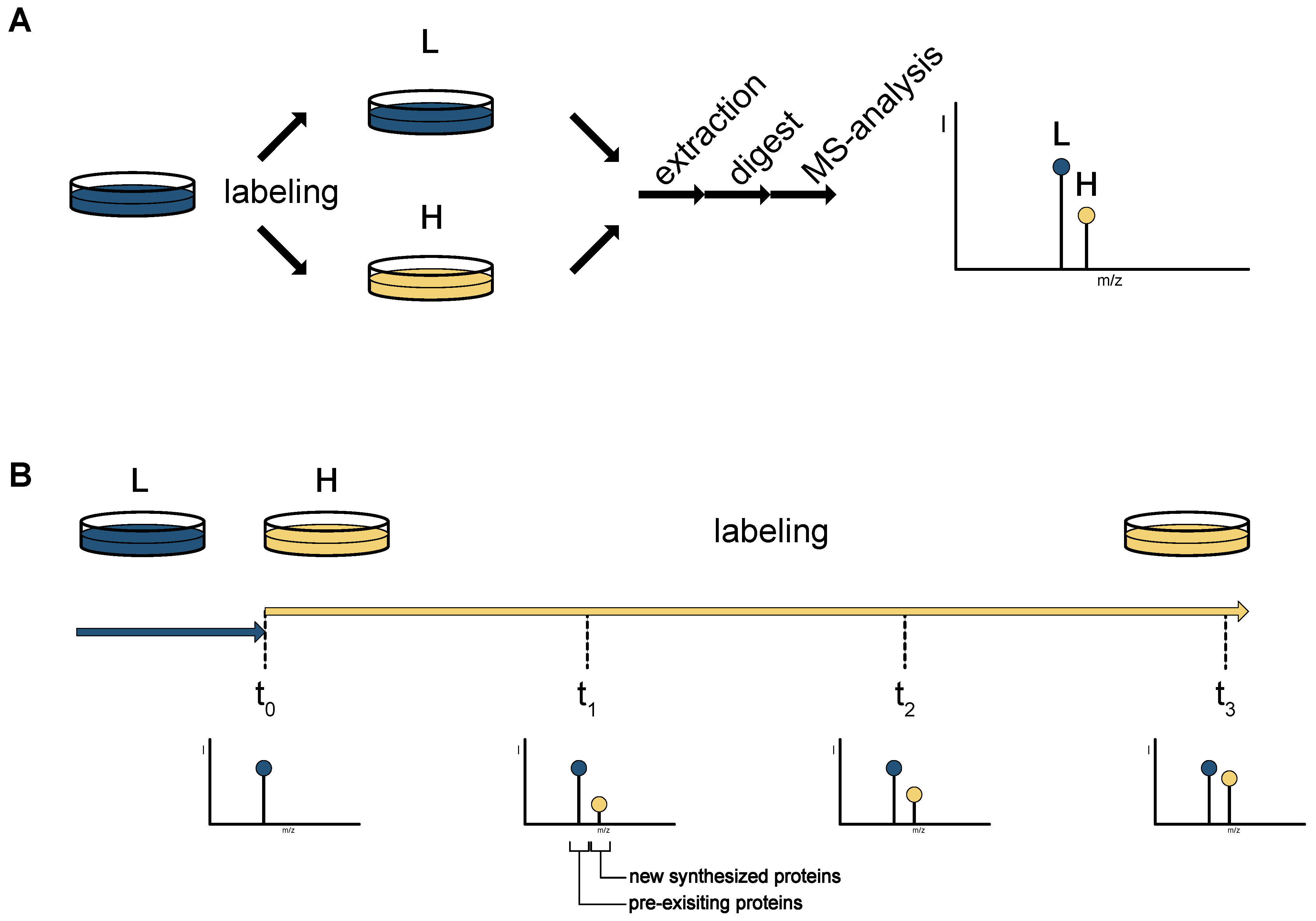
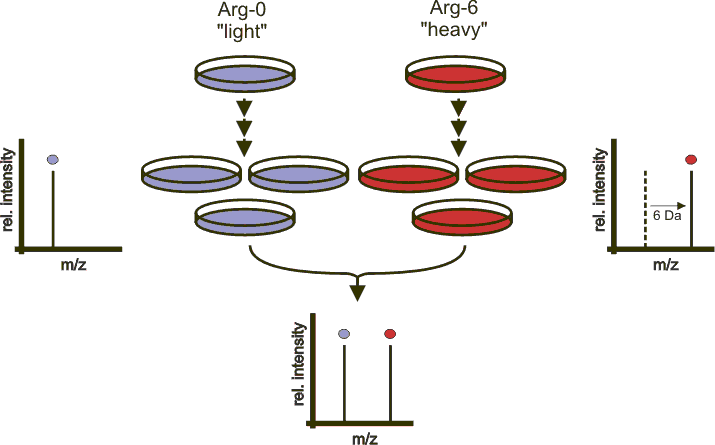
Quantitative proteomics using SILAC: Principles, applications, and developments - Chen - 2015 - PROTEOMICS - Wiley Online Library
PLOS ONE: Semi-quantitative proteomics of mammalian cells upon short-term exposure to non-ionizing electromagnetic fields
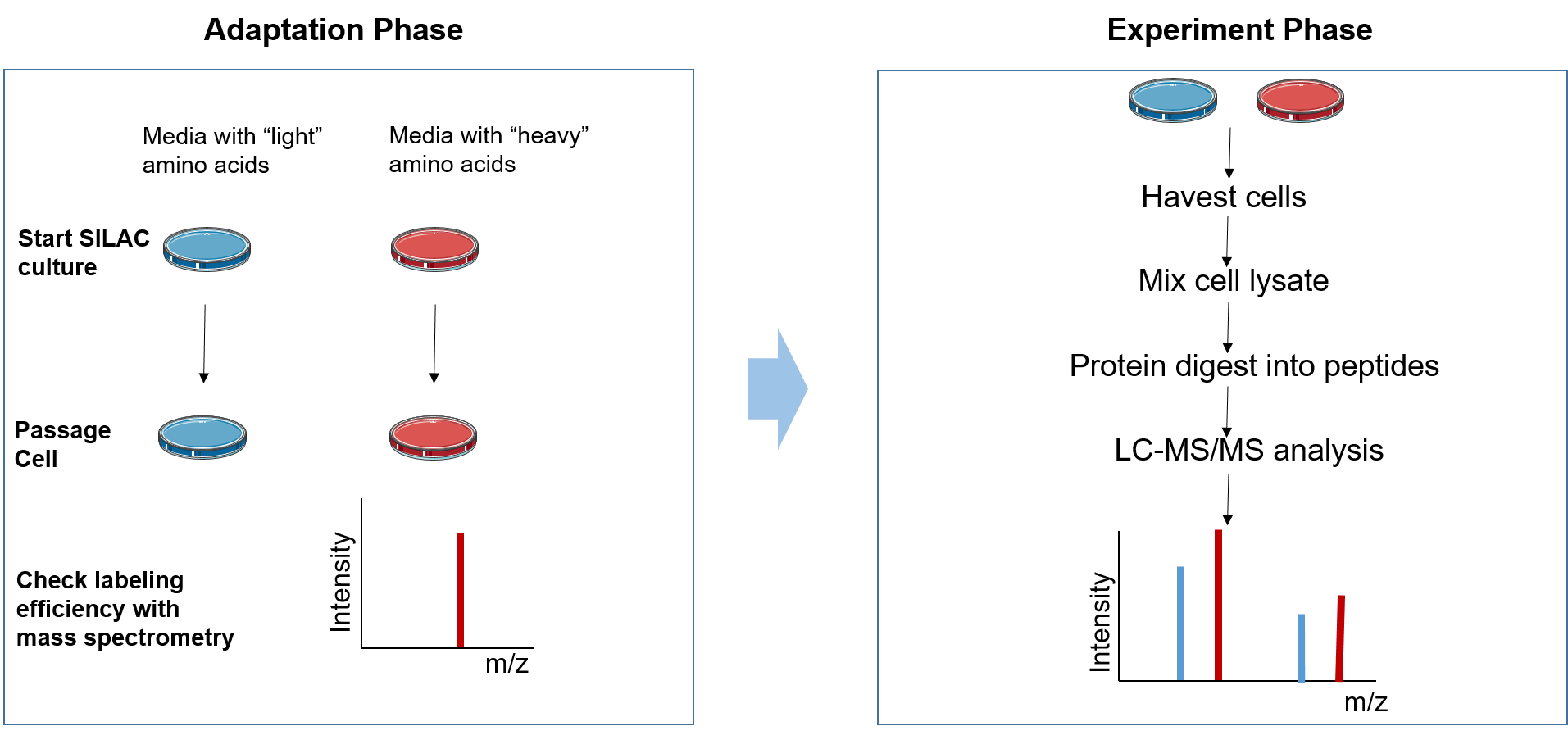
Stable Isotope Labeling Using Amino Acids in Cell Culture (SILAC): Principles, Workflow, and Applications – Creative Proteomics Blog
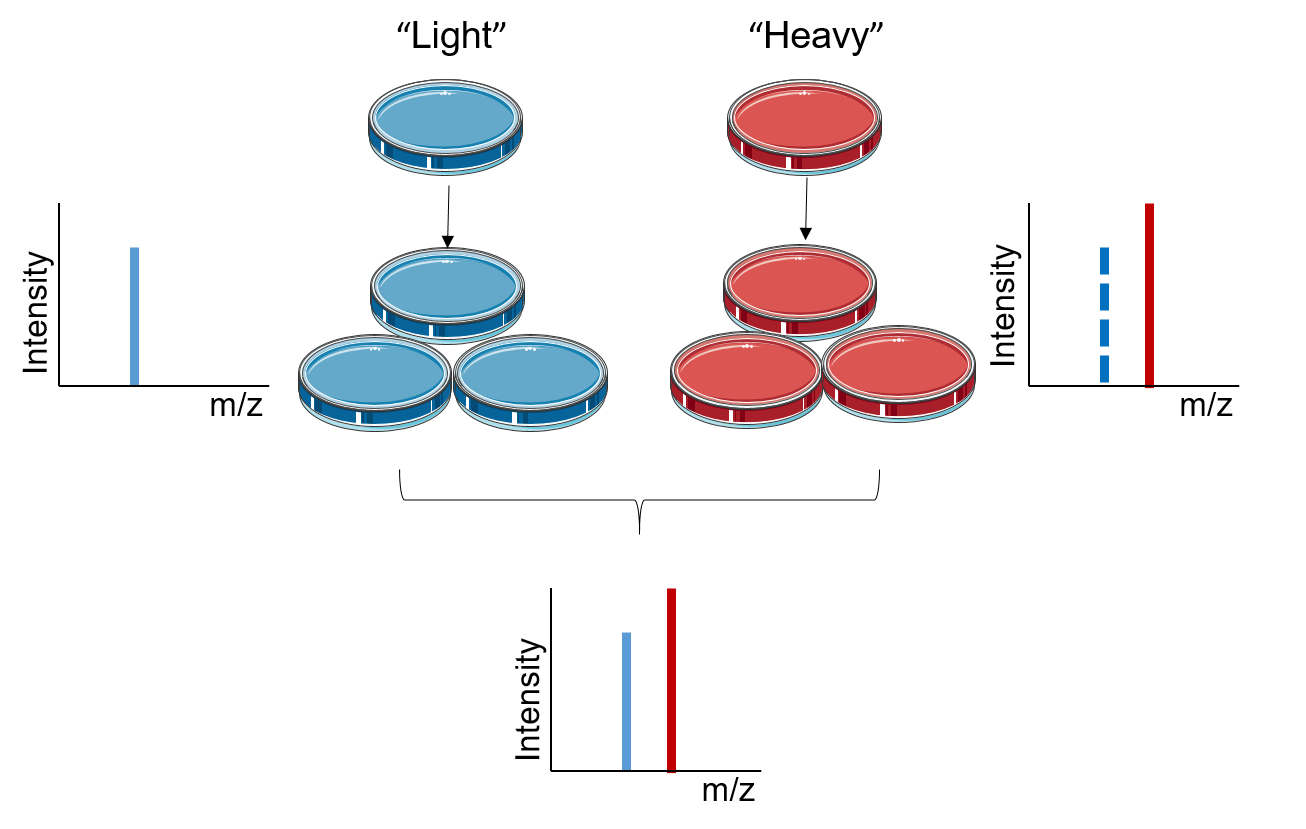
Stable Isotope Labeling Using Amino Acids in Cell Culture (SILAC): Principles, Workflow, and Applications – Creative Proteomics Blog

Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics* - Molecular & Cellular Proteomics

An Enhanced In Vivo Stable Isotope Labeling by Amino Acids in Cell Culture ( SILAC) Model for Quantification of Drug Metabolism Enzymes * - Molecular & Cellular Proteomics

SILAC Mouse for Quantitative Proteomics Uncovers Kindlin-3 as an Essential Factor for Red Blood Cell Function: Cell

JCI Insight - Deep phenotyping of human induced pluripotent stem cell–derived atrial and ventricular cardiomyocytes

Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics | Nature Protocols

A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) | Semantic Scholar

Determination of an Angiotensin II-regulated Proteome in Primary Human Kidney Cells by Stable Isotope Labeling of Amino Acids in Cell Culture ( SILAC) - Journal of Biological Chemistry

Amino acid based labeling techniques. (A) SILAC labeling. Proteins are... | Download Scientific Diagram
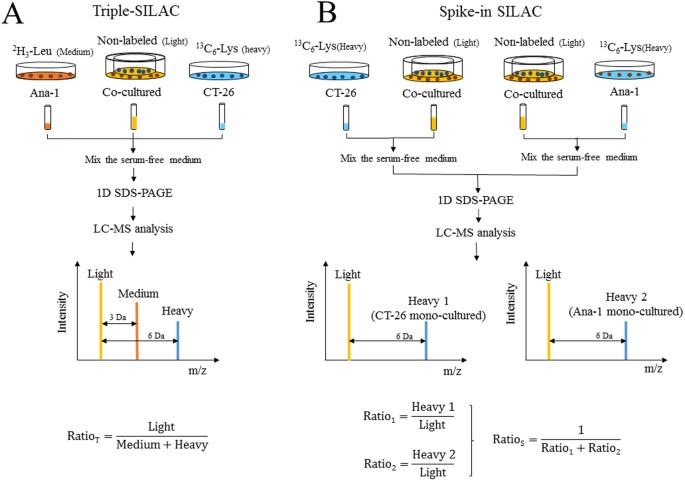
SILAC–based quantitative MS approach for real-time recording protein-mediated cell-cell interactions | Scientific Reports

Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) for Studying Dynamics of Protein Abundance and Posttranslational Modifications

Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) for Quantitative Proteomics | SpringerLink